Photon Classification
This Jupyter notebook can be downloaded here.
Photon Classification
Photons in this analysis are identified using photon ID variables. However, other particles can also pass this ID. For example, a boosted jet can decay into two photons in such a way that they overlap within the resolution of the detector, mimicking a single photon signature and causing the jet to be misidentified as a (“fake”) photon.
We want to classify all particles passing the photon ID as either real or fake:
- Real photons ($y=0$)
- Fake photons ($y=1$)
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.lines import Line2D
import seaborn as sns
#from scipy import stats
import sklearn
import sklearn.linear_model
from sklearn import preprocessing
from sklearn.neural_network import MLPClassifier
from keras import Sequential
from keras.layers import Dense
ROOT Data Format
The data has been initially processed using the CERN ROOT data analysis frame work and is stored in trees using its TTree() data structure. Observe the following tree->branch->leaf format.
This will be converted into a pandas DataFrame using root_pandas.
#import ROOT
from root_pandas import read_root
Welcome to JupyROOT 6.16/00
Simulated Data
The dataset has been constructed using Monte Carlo simulation. The particle collisions and interactions have been generated using PYTHIA 8 and simulated through the CMS detector using Geant4. This simulation allows us to label all particles passing the photon ID as either real or fake. Note, for actual detector data we do not have this luxury.
fQCD1 = '~/diphoton_fake_rate_closure_test_QCD_Pt_1000to1400_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD2 = '~/diphoton_fake_rate_closure_test_QCD_Pt_10to15_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD3 = '~/diphoton_fake_rate_closure_test_QCD_Pt_120to170_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD4 = '~/diphoton_fake_rate_closure_test_QCD_Pt_1400to1800_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD5 = '~/diphoton_fake_rate_closure_test_QCD_Pt_15to30_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD6 = '~/diphoton_fake_rate_closure_test_QCD_Pt_170to300_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD7 = '~/diphoton_fake_rate_closure_test_QCD_Pt_1800to2400_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD8 = '~/diphoton_fake_rate_closure_test_QCD_Pt_2400to3200_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD9 = '~/diphoton_fake_rate_closure_test_QCD_Pt_300to470_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD10 = '~/diphoton_fake_rate_closure_test_QCD_Pt_30to50_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD11 = '~/diphoton_fake_rate_closure_test_QCD_Pt_3200toInf_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD12 = '~/diphoton_fake_rate_closure_test_QCD_Pt_470to600_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD13 = '~/diphoton_fake_rate_closure_test_QCD_Pt_50to80_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD14 = '~/diphoton_fake_rate_closure_test_QCD_Pt_5to10_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD15 = '~/diphoton_fake_rate_closure_test_QCD_Pt_600to800_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD16 = '~/diphoton_fake_rate_closure_test_QCD_Pt_800to1000_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
fQCD17 = '~/diphoton_fake_rate_closure_test_QCD_Pt_80to120_TuneCUETP8M1_13TeV_pythia8_76X_MiniAOD_merged.root'
files_QCD = [fQCD1,fQCD2,fQCD3,fQCD4,fQCD5,fQCD6,fQCD7,fQCD8,fQCD9,fQCD10,fQCD11,fQCD12,fQCD13,fQCD14,fQCD15,fQCD16,fQCD17]
fGJets1 = '~/diphoton_fake_rate_closure_test_GJets_HT-100To200_TuneCUETP8M1_13TeV-madgraphMLM-pythia8_76X_MiniAOD_merged.root'
fGJets2 = '~/diphoton_fake_rate_closure_test_GJets_HT-200To400_TuneCUETP8M1_13TeV-madgraphMLM-pythia8_76X_MiniAOD_merged.root'
fGJets3 = '~/diphoton_fake_rate_closure_test_GJets_HT-400To600_TuneCUETP8M1_13TeV-madgraphMLM-pythia8_76X_MiniAOD_merged.root'
fGJets4 = '~/diphoton_fake_rate_closure_test_GJets_HT-40To100_TuneCUETP8M1_13TeV-madgraphMLM-pythia8_76X_MiniAOD_merged.root'
fGJets5 = '~/diphoton_fake_rate_closure_test_GJets_HT-600ToInf_TuneCUETP8M1_13TeV-madgraphMLM-pythia8_76X_MiniAOD_merged.root'
files_GJets = [fGJets1,fGJets2,fGJets3,fGJets4,fGJets5]
df = read_root(files_QCD,'diphoton/fTreeFake',columns=['Photon','PhotonGenMatch','Event'])
df.head()
| Event_run | Event_LS | Event_evnum | Event_processid | Event_bx | Event_orbit | Event_ptHat | Event_alphaqcd | Event_alphaqed | Event_qscale | ... | Photon_passCorPhoIso | Photon_passSieie | Photon_passHighPtID | Photon_passChIsoDenom | Photon_passCorPhoIsoDenom | Photon_isFakeable | Photon_isNumeratorObjCand | Photon_isDenominatorObj | Photon_isSaturated | Photon_isMCTruthFake | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 500 | 66374 | 113 | -1 | -1 | 1026.243164 | 0.095640 | 0.008038 | 1026.243164 | ... | True | True | True | True | True | False | True | False | False | False |
| 1 | 1 | 588 | 78098 | 114 | -1 | -1 | 1002.348999 | 0.095880 | 0.008036 | 1002.349060 | ... | True | True | False | True | True | False | True | True | False | False |
| 2 | 1 | 648 | 86108 | 113 | -1 | -1 | 1142.139282 | 0.094561 | 0.008048 | 1142.139282 | ... | True | True | True | True | True | False | True | False | False | False |
| 3 | 1 | 648 | 86140 | 113 | -1 | -1 | 1063.714844 | 0.095275 | 0.008041 | 1063.714844 | ... | True | True | True | True | True | False | True | False | False | False |
| 4 | 1 | 681 | 90441 | 114 | -1 | -1 | 1008.274658 | 0.095820 | 0.008036 | 1008.274658 | ... | True | True | False | True | True | False | True | False | False | False |
5 rows × 100 columns
df.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 443783 entries, 0 to 443782
Data columns (total 100 columns):
Event_run 443783 non-null int64
Event_LS 443783 non-null int64
Event_evnum 443783 non-null int64
Event_processid 443783 non-null int64
Event_bx 443783 non-null int64
Event_orbit 443783 non-null int64
Event_ptHat 443783 non-null float32
Event_alphaqcd 443783 non-null float32
Event_alphaqed 443783 non-null float32
Event_qscale 443783 non-null float32
Event_x1 443783 non-null float32
Event_x2 443783 non-null float32
Event_pdf1 443783 non-null float32
Event_pdf2 443783 non-null float32
Event_weight0 443783 non-null float32
Event_weight 443783 non-null float32
Event_weightPuUp 443783 non-null float32
Event_weightPu 443783 non-null float32
Event_weightPuDown 443783 non-null float32
Event_weightLumi 443783 non-null float32
Event_weightAll 443783 non-null float32
Event_interactingParton1PdgId 443783 non-null int32
Event_interactingParton2PdgId 443783 non-null int32
Event_pdf_id1 443783 non-null int32
Event_pdf_id2 443783 non-null int32
Event_npv_true 443783 non-null int32
Event_beamHaloIDLoose 443783 non-null bool
Event_beamHaloIDTight 443783 non-null bool
Event_beamHaloIDTight2015 443783 non-null bool
PhotonGenMatch_pt 443783 non-null float64
PhotonGenMatch_eta 443783 non-null float64
PhotonGenMatch_phi 443783 non-null float64
PhotonGenMatch_deltaR_match 443783 non-null float64
PhotonGenMatch_deltaR_matchDau 443783 non-null float64
PhotonGenMatch_ptDiff_match 443783 non-null float64
PhotonGenMatch_matchCategory 443783 non-null int32
PhotonGenMatch_matchType 443783 non-null int32
PhotonGenMatch_nPhotonMotherDaughters 443783 non-null int32
PhotonGenMatch_status 443783 non-null int32
PhotonGenMatch_motherStatus 443783 non-null int32
PhotonGenMatch_grandmotherStatus 443783 non-null int32
PhotonGenMatch_pdgId 443783 non-null int32
PhotonGenMatch_motherPdgId 443783 non-null int32
PhotonGenMatch_grandmotherPdgId 443783 non-null int32
Photon_pt 443783 non-null float64
Photon_eta 443783 non-null float64
Photon_phi 443783 non-null float64
Photon_scEta 443783 non-null float64
Photon_scPhi 443783 non-null float64
Photon_rho 443783 non-null float64
Photon_chargedHadIso03 443783 non-null float64
Photon_neutralHadIso03 443783 non-null float64
Photon_photonIso03 443783 non-null float64
Photon_rhoCorChargedHadIso03 443783 non-null float64
Photon_rhoCorNeutralHadIso03 443783 non-null float64
Photon_rhoCorPhotonIso03 443783 non-null float64
Photon_corPhotonIso03 443783 non-null float64
Photon_hadTowerOverEm 443783 non-null float64
Photon_hadronicOverEm 443783 non-null float64
Photon_r9 443783 non-null float64
Photon_r9_5x5 443783 non-null float64
Photon_sigmaIetaIeta 443783 non-null float64
Photon_sigmaIetaIeta5x5 443783 non-null float64
Photon_sigmaEtaEta 443783 non-null float64
Photon_sigmaIphiIphi 443783 non-null float64
Photon_sigmaIphiIphi5x5 443783 non-null float64
Photon_sigmaIetaIphi 443783 non-null float64
Photon_sigmaIetaIphi5x5 443783 non-null float64
Photon_maxEnergyXtal 443783 non-null float64
Photon_iEta 443783 non-null float64
Photon_iPhi 443783 non-null float64
Photon_alphaHighPtID 443783 non-null float64
Photon_kappaHighPtID 443783 non-null float64
Photon_phoEAHighPtID 443783 non-null float64
Photon_chEAegmID 443783 non-null float64
Photon_nhEAegmID 443783 non-null float64
Photon_phoEAegmID 443783 non-null float64
Photon_passEGMLooseID 443783 non-null bool
Photon_passEGMMediumID 443783 non-null bool
Photon_passEGMTightID 443783 non-null bool
Photon_isEB 443783 non-null bool
Photon_isEE 443783 non-null bool
Photon_isEBEtaGap 443783 non-null bool
Photon_isEBPhiGap 443783 non-null bool
Photon_isEERingGap 443783 non-null bool
Photon_isEEDeeGap 443783 non-null bool
Photon_isEBEEGap 443783 non-null bool
Photon_passElectronVeto 443783 non-null bool
Photon_passHTowOverE 443783 non-null bool
Photon_passChIso 443783 non-null bool
Photon_passCorPhoIso 443783 non-null bool
Photon_passSieie 443783 non-null bool
Photon_passHighPtID 443783 non-null bool
Photon_passChIsoDenom 443783 non-null bool
Photon_passCorPhoIsoDenom 443783 non-null bool
Photon_isFakeable 443783 non-null bool
Photon_isNumeratorObjCand 443783 non-null bool
Photon_isDenominatorObj 443783 non-null bool
Photon_isSaturated 443783 non-null bool
Photon_isMCTruthFake 443783 non-null bool
dtypes: bool(26), float32(15), float64(39), int32(14), int64(6)
memory usage: 212.5 MB
- By construction, there are no NULL values. Let’s sanity check this.
- Initially, there are $m = 443,783$ examples. Some preprocessing is in order though.
#df.describe(include='all')
df.isnull().sum().sum()
0
len(df)
443783
Data Preprocessing
We apply the following pre-selection on all examples:
- Pass photon ID
- Located inside the EB or EE detectors (these are the CMS ECAL barrel and endcap detectors, respectively)
- Are high energy, above 50 GeV
- Have small shower shapes along the $\phi$-direction (this eliminates a certain type of fakes produced from another physical source)
Now, $m = 156,202$
df = df[df['Photon_passHighPtID'] == 1]
df = df[(df['Photon_isEB']==True) | (df['Photon_isEE']==True)]
df = df[(df['Photon_pt'] > 50.0) & (df['Photon_sigmaIphiIphi5x5'] > 0.009)]
len(df)
156202
Data Visualization
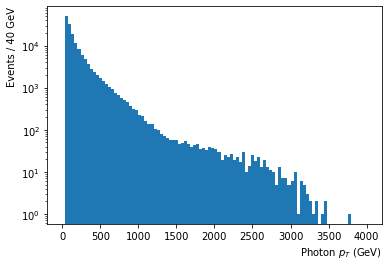
- Photon $p_T$ shows most photons fall towards the lower side of the energy spectrum
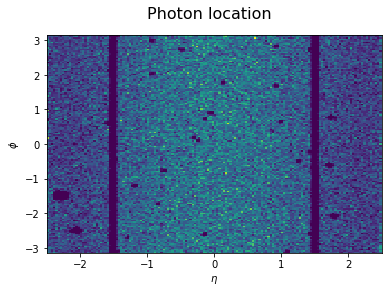
- Photon $\eta$ vs. $\phi$ shows where the photons are located inside the ECAL (with the EB and EE separated by a gap on both sides)
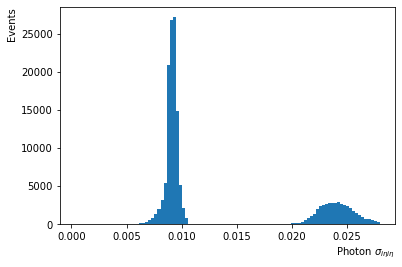
- Photon $\sigma_{i\eta i\eta}$ shows the width of the photon shower along the $\eta$-direction (with different distributions in the EB and EE regions)
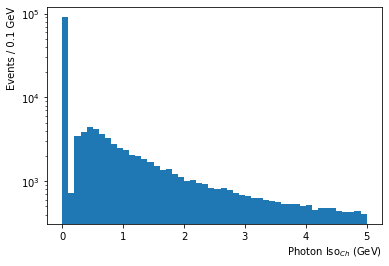
- Photon Iso$_{Ch}$ shows that most photons have little activity around them and are well isolated
n_bins_pt = 100;
#(max(df['Photon_pt'])-min(df['Photon_pt']))/60
#bins=np.arange(min(data), max(data) + binwidth, binwidth)
df['Photon_pt'].plot.hist(bins=n_bins_pt,range=(0,4000),log=True); #weights=df['Event_weightAll']
plt.xlabel('Photon $p_T$ (GeV)', horizontalalignment='right', x=1.0);
plt.ylabel('Events / '+str(int(4000/n_bins_pt))+' GeV', horizontalalignment='right', y=1.0);
#df.plot.scatter('Photon_scEta', 'Photon_scPhi');
plt.hist2d(df['Photon_scEta'],df['Photon_scPhi'],bins=150); #weights=df['Event_weightAll']
plt.suptitle('Photon location',fontsize=16)
plt.xlabel('$\eta$');
plt.ylabel('$\phi$');
df['Photon_sigmaIetaIeta5x5'].plot.hist(bins=100,log=False); #weights=df['Event_weightAll']
plt.xlabel('Photon $\sigma_{i\eta i\eta}$', horizontalalignment='right', x=1.0);
plt.ylabel('Events', horizontalalignment='right', y=1.0);
df['Photon_chargedHadIso03'].plot.hist(bins=50,log=True); #weights=df['Event_weightAll']
plt.xlabel('Photon Iso$_{Ch}$ (GeV)', horizontalalignment='right', x=1.0);
plt.ylabel('Events / '+str(5/50)+' GeV', horizontalalignment='right', y=1.0);
Create labels
Let’s encode the fake criteria into a single label:
- Real photons ($y=0$)
- Fake photons ($y=1$)
Store this as a NumPy array Y_train.
def is_fake(row):
if (row['Photon_passHighPtID'] == 1) and ((row['PhotonGenMatch_matchCategory'] == 1) or (row['PhotonGenMatch_matchCategory'] == 2)) and (row['PhotonGenMatch_matchType'] == 1):
val = 1
else:
val = 0
return val
df['fake'] = df.apply(is_fake, axis=1)
#df[['PhotonGenMatch_matchCategory','PhotonGenMatch_matchType','fake']][df['fake']==1]
df['fake'].value_counts()
0 138435
1 17767
Name: fake, dtype: int64
Y = df['fake'].to_numpy()
Y.shape
#Y_train = Y.reshape(Y.shape[0],1)
#Y_train.shape
(156202,)
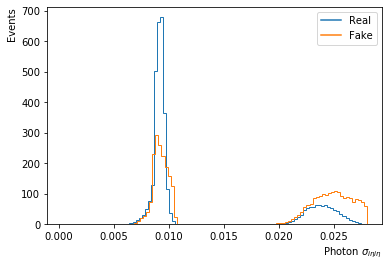
fig = plt.figure()
ax = fig.add_subplot(111)
df['Photon_sigmaIetaIeta5x5'][df['fake']==0].plot.hist(bins=100,density=True,log=False,label='Real',histtype='step'); # weights=df['Event_weightAll'][df['fake']==0]
df['Photon_sigmaIetaIeta5x5'][df['fake']==1].plot.hist(bins=100,density=True,log=False,label='Fake',histtype='step'); # weights=df['Event_weightAll'][df['fake']==1]
plt.xlabel('Photon $\sigma_{i\eta i\eta}$', horizontalalignment='right', x=1.0);
plt.ylabel('Events', horizontalalignment='right', y=1.0);
plt.legend(loc='upper right');
handles, labels = ax.get_legend_handles_labels()
new_handles = [Line2D([], [], c=h.get_edgecolor()) for h in handles]
plt.legend(handles=new_handles, labels=labels)
plt.show()
Prepare features
Organize desired features into NumPy array X_train. Normalize features by mean and standardize by the standard deviation.
features = ['Photon_pt','Photon_eta','Photon_phi','Photon_iEta','Photon_iPhi','Photon_rho','Photon_chargedHadIso03','Photon_neutralHadIso03','Photon_photonIso03','Photon_hadTowerOverEm','Photon_r9_5x5','Photon_sigmaIetaIeta5x5','Photon_sigmaIphiIphi5x5','Photon_sigmaIetaIphi5x5','Photon_maxEnergyXtal']
X = df[features].to_numpy()
X.shape
(156202, 15)
scaler = preprocessing.StandardScaler().fit(X)
X_scaled = scaler.transform(X)
X_scaled.shape
(156202, 15)
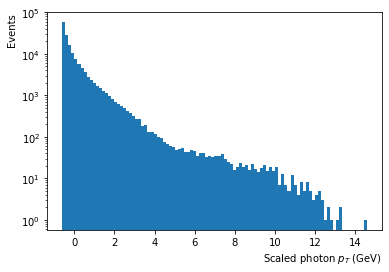
plt.hist(x=X_scaled[:,0], bins=100,log=True);
plt.xlabel('Scaled photon $p_T$ (GeV)', horizontalalignment='right', x=1.0);
plt.ylabel('Events', horizontalalignment='right', y=1.0);
Feature Correlation
- The strongest correlations are among the angular coordinates, shower shapes along the angular coordinates, and among the different energy variables
corr = df[features].corr()
sns.heatmap(corr);

Split dataset
Let’s divided the dataset into training and testing datasets.
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(X_scaled, Y, test_size=0.3)
print ("Number of total examples: " + str(X.shape[0]))
print ("Number of training examples: " + str(X_train.shape[0]))
print ("Number of testing examples: " + str(X_test.shape[0]))
print ("X_train shape: " + str(X_train.shape))
print ("Y_train shape: " + str(Y_train.shape))
print ("X_test shape: " + str(X_test.shape))
print ("Y_test shape: " + str(Y_test.shape))
Number of total examples: 156202
Number of training examples: 109341
Number of testing examples: 46861
X_train shape: (109341, 15)
Y_train shape: (109341,)
X_test shape: (46861, 15)
Y_test shape: (46861,)
Simple Logistic Regression Classifier
We train a simple linear classifier using logistic regression to get a baseline for performance on this dataset.
clf = sklearn.linear_model.LogisticRegressionCV();
clf.fit(X_train, Y_train);
/Users/abuccilli/anaconda3/envs/photon-classification-keras/lib/python3.7/site-packages/sklearn/model_selection/_split.py:1978: FutureWarning: The default value of cv will change from 3 to 5 in version 0.22. Specify it explicitly to silence this warning.
warnings.warn(CV_WARNING, FutureWarning)
LR_predictions_train = clf.predict(X_train)
print ('Accuracy of logistic regression: %d' % float((np.dot(Y_train,LR_predictions_train) + np.dot(1-Y_train,1-LR_predictions_train))/float(Y_train.size)*100) +'% ' + "(training set)")
Accuracy of logistic regression: 92% (training set)
LR_predictions_test = clf.predict(X_test)
print ('Accuracy of logistic regression: %d' % float((np.dot(Y_test,LR_predictions_test) + np.dot(1-Y_test,1-LR_predictions_test))/float(Y_test.size)*100) +'% ' + "(test set)")
Accuracy of logistic regression: 92% (test set)
- The accuracy is promising; however, the classes are skewed so let’s check the F1 score.
sklearn.metrics.f1_score(Y_test, LR_predictions_test)
0.5984450034301395
np.dot(Y_test,LR_predictions_test)/sum(Y_test==1)*100.
48.21296978629329
np.dot(1-Y_test,1-LR_predictions_test)/sum(Y_test==0)*100.
98.30811189148746
Nonlinear classifier
We use a 4-layer deep neural network as our classifier, comprising 3 hidden layers. L2 regularization is used.
clf_NN = MLPClassifier(solver='lbfgs', alpha=1e-2, hidden_layer_sizes=(10,10,4), max_iter=1500, learning_rate_init=0.0001, random_state=1)
clf_NN.fit(X_train,Y_train)
MLPClassifier(activation='relu', alpha=0.01, batch_size='auto', beta_1=0.9,
beta_2=0.999, early_stopping=False, epsilon=1e-08,
hidden_layer_sizes=(10, 10, 4), learning_rate='constant',
learning_rate_init=0.0001, max_iter=1500, momentum=0.9,
n_iter_no_change=10, nesterovs_momentum=True, power_t=0.5,
random_state=1, shuffle=True, solver='lbfgs', tol=0.0001,
validation_fraction=0.1, verbose=False, warm_start=False)
NN_predictions_train = clf_NN.predict(X_train)
print ('Accuracy of NN: %d' % float((np.dot(Y_train,NN_predictions_train) + np.dot(1-Y_train,1-NN_predictions_train))/float(Y_train.size)*100) +'% ' + "(training set)")
Accuracy of NN: 94% (training set)
NN_predictions_test = clf_NN.predict(X_test)
print ('Accuracy of NN: %d' % float((np.dot(Y_test,NN_predictions_test) + np.dot(1-Y_test,1-NN_predictions_test))/float(Y_test.size)*100) +'% ' + "(test set)")
Accuracy of NN: 94% (test set)
sklearn.metrics.f1_score(Y_test, NN_predictions_test)
0.7239679605668514
np.dot(Y_test,NN_predictions_test)/sum(Y_test==1)*100.
65.16916250693289
np.dot(1-Y_test,1-NN_predictions_test)/sum(Y_test==0)*100.
98.06040721798706
DNN with Keras
classifier = Sequential()
classifier.add(Dense(10, activation='relu', kernel_initializer='normal', input_dim=len(features)))
classifier.add(Dense(10, activation='relu', kernel_initializer='normal'))
classifier.add(Dense(4, activation='relu', kernel_initializer='normal'))
classifier.add(Dense(1, activation='sigmoid', kernel_initializer='normal'))
classifier.compile(optimizer ='adam',loss='binary_crossentropy', metrics =['accuracy'])
classifier.fit(X_train,Y_train, batch_size=100, epochs=10)
Epoch 1/10
109341/109341 [==============================] - 2s 15us/step - loss: 0.2623 - acc: 0.8885
Epoch 2/10
109341/109341 [==============================] - 1s 11us/step - loss: 0.2006 - acc: 0.9317
Epoch 3/10
109341/109341 [==============================] - 1s 11us/step - loss: 0.1914 - acc: 0.9334
Epoch 4/10
109341/109341 [==============================] - 1s 11us/step - loss: 0.1856 - acc: 0.9346
Epoch 5/10
109341/109341 [==============================] - 1s 12us/step - loss: 0.1825 - acc: 0.9351
Epoch 6/10
109341/109341 [==============================] - 1s 12us/step - loss: 0.1805 - acc: 0.9355
Epoch 7/10
109341/109341 [==============================] - 1s 11us/step - loss: 0.1792 - acc: 0.9358
Epoch 8/10
109341/109341 [==============================] - 1s 11us/step - loss: 0.1782 - acc: 0.9361
Epoch 9/10
109341/109341 [==============================] - 1s 11us/step - loss: 0.1772 - acc: 0.9366
Epoch 10/10
109341/109341 [==============================] - 1s 12us/step - loss: 0.1766 - acc: 0.9366
<keras.callbacks.History at 0x13a63f320>
classifier.evaluate(X_train, Y_train)
109341/109341 [==============================] - 1s 11us/step
[0.17581189132354602, 0.9365014038709693]
Y_pred_keras = classifier.predict(X_test)
Y_pred_keras = (Y_pred_keras > 0.5)
print ('Accuracy of NN: %d' % float((np.dot(Y_test,Y_pred_keras) + np.dot(1-Y_test,1-Y_pred_keras))/float(Y_test.size)*100) +'% ' + "(test set)")
Accuracy of NN: 93% (test set)
sklearn.metrics.f1_score(Y_test, Y_pred_keras)
0.6714394185920382
np.squeeze(np.dot(Y_test,Y_pred_keras)/sum(Y_test==1))*100.
56.54016212232867
np.squeeze(np.dot(1-Y_test,1-Y_pred_keras)/sum(Y_test==0))*100.
98.33224724253614
Test on Production Data
We further check our classifier’s performance by checking it on other data. First, we need to preprocess our features. This dataset is heavily skewed between classes.
df_test = read_root(fGJets2,'diphoton/fTreeFake',columns=['Photon','PhotonGenMatch','Event'])
df_test = df_test[df_test['Photon_passHighPtID'] == 1]
df_test = df_test[(df_test['Photon_isEB']==True) | (df_test['Photon_isEE']==True)]
df_test = df_test[(df_test['Photon_pt'] > 50.0) & (df_test['Photon_sigmaIphiIphi5x5'] > 0.009)]
len(df_test)
1869221
df_test['fake'] = df_test.apply(is_fake, axis=1)
df_test['fake'].value_counts()
0 1862201
1 7020
Name: fake, dtype: int64
Y_prod = df_test['fake'].to_numpy()
X_prod = df_test[features].to_numpy()
X_prod_scaled = scaler.transform(X_prod)
NN_predictions_prod = clf_NN.predict(X_prod_scaled)
print ('Accuracy of NN: %d' % float((np.dot(Y_prod,NN_predictions_prod) + np.dot(1-Y_prod,1-NN_predictions_prod))/float(Y_prod.size)*100) +'% ' + "(production test set)")
Accuracy of NN: 97% (production test set)
sklearn.metrics.f1_score(Y_prod, NN_predictions_prod)
0.15087430193170373
np.dot(Y_prod,NN_predictions_prod)/sum(Y_prod==1)*100.
58.68945868945868
np.dot(1-Y_prod,1-NN_predictions_prod)/sum(Y_prod==0)*100.
97.66539702212597
